Using bond valence sums in Artemis
The concept of a bond in inorganic or crystal chemistry is a bit
ambiguous. In a seminal pair of papers,
D. Altermatt and I. D. Brown, Acta Cryst. B, 41,
(1985) p. 240-244 (DOI: 10.1107/S0108768185002051)
and
I. D. Brown and D. Altermatt, Acta Cryst. B, 41,
(1985) p. 244-247 (DOI: 10.1107/S0108768185002063)
Brown and Altermatt proposed this definition
“All neighbouring cation-anion distances are
considered to be bonds although not all of equal strength.”
In this model, each bond between atoms i and j has a number –
the bond valence – sij which is inversely proportional to
bond distance. The bond valence is defined as
sij=exp((R0,ij-Rij)/B), where Rij is the contact
distance and R0,ij and B are empirically determined parameters.
They searched through the Inorganic Crystal Structures Database to
determine empirical values for sij for over 150 cation/anion
pairs. Other authors have supplemented this work with additional
anion/cation pairs. Most of these are for common cations, such as
oxygen, nitrogen, or sulfur. Interestingly, B is nearly constant
across all bonds and equal to 0.37. For some anions such as K and U,
the value of B can be as high as about 0.6. R0,ij depends on the
contact pair and has been tabulated along with B in a database.
The bond valance sum, then, is the sum of sij over all pairs in a
coordination shell: V=Σsij. The bond valence sum V should
be equal to the formal valence of the absorber cation. This provides
a way of relating coordination number, bond distance, and formal
valence in a way that is useful and directly applicable to EXAFS
analysis.
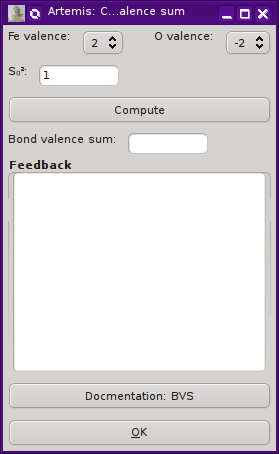
Computing a bond valence sum from a fit
ARTEMIS provides a tool for computing a bond valence sum from a
list of paths included in a fit. A set of paths to be included in the
sum can be marked in a path list. From the Actions menu, selecting
“Compute bond valence sum” will display the
dialog on the right.
Some care is taken to verify that your selection of paths is
sensible. ARTEMIS will notice if you have marked multiple
scattering paths or have marked paths with absorber/scatterer pairs
that are not in the bond valence database. Although it will proceed
with a calculation, ARTEMIS will warn you if it seems as though
you have included paths that do not seem to be a part of the first
coordination shell.
ARTEMIS also tries to make good guesses about the formal valences
of the absorber and scatterer, althoguh it will often guess wrongly.
It is, therefore, essential that you set the valences correctly using
the choice menus at the top of the bond valence dialog. It is much
more likely that the absorber valence will be guessed incorrectly.
You will notice that one of the valence options for many absorber
species is “9”, an obviously wrong value of
valence. The bond valence database says “Bond
valence parameters for atoms whose oxidation state is given as 9 do
not have an oxidation state specified in the original citation. They
may apply to a particular, but unspecified, oxidation state or they
may be intended to apply to all oxidation states.”
In order to make the bond valence summation, the degeneracy of each
path included in the sum must be multiplied by its evaluation of
sij (which also uses the evaluation of R=R0+ΔR as the
value of Rij). Because path degeneracy might need to consider
quite complicated parameterization of the S²₀ path parameter as
well as the N path parameter, ARTEMIS will multiply the
evaluations of the N and S²₀ path parameters together to use as
the evaluation of degeneracy in the summmation. It is up to you, the
user, to supply a value for the actual amplitude reduction factor,
S²₀ to be divided out of the summation.
Pressing the “Compute” button will make the
bond valence sum, reporting its value in the text box. Any
feedback will be written in the larger text control. For a successful
calculation, the values of Rij and B obtained from the database
will be displayed. Any warnings about the path selection will be
printed in the feedback box in bold red text.
Using a bond valance sum as a restraint
The bond valence sum can be used a restraint on a fit. That is, the
relationship between formal valence, coordination number, and bond
distance can be used as prior knowledge guiding the fit. If the
absorber/scatterer pair are in the bond valence database, values for
R0,ij, B, and the formal valence of the obsorber can be defined as
set parameters. The bond valence sum is expressed as a def
parameter. Finally, the difference between the bond valence sum and
the formal valence are expressed as a restrain parameter. These
are shown below for the Fe-O bond in FeO. In FeO the iron atom is of
valence 2+ and the oxygen is 2-.
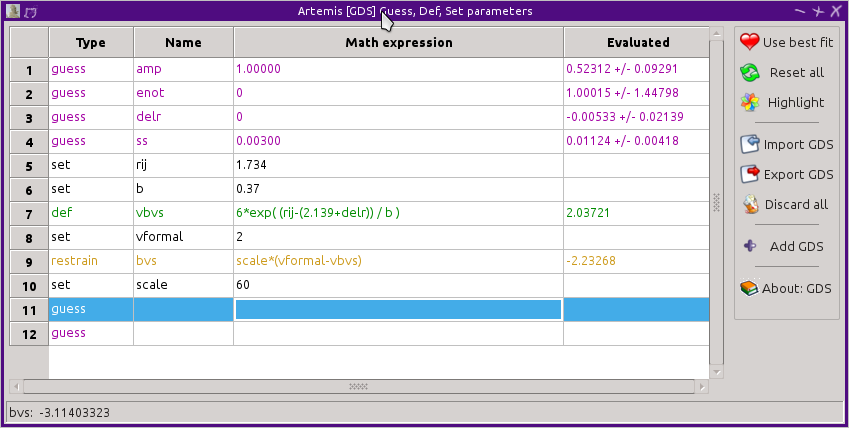
When the fit is evaluated, the restrain parameter will be added in
quadrature to the evaluation of χ². This sum will be
minimized in the fit. In a fit to FeO, the coordination number is
fixed to 6, the value known from cyrstallography. By using this
restraint, the value of ΔR will be encouraged to assume a value
that results in a bond valence sum of 2. By increasing the value of
the scale parameter, the strength of the restraint is increased. For
a very large value of scale, ΔR will constrained to a value that
forces the bond valence sum to 2. For a very small value of scale,
the restraint will be weak and ΔR will be given more freedom to
deviate from a value that casues a bond valence sum of 2.
This example shows the simplest case of a single scattering path
contributing to the bond valence sum. The math expressions to
establish the restraint would be more complicated for a more
disrodered first shell, but those math expressions would follow the
same pattern as this example.
Using a bond valance sum as an after parameter
The last ARTEMIS trick related to evaluations of bond valence sums
is to use an after parameter to record the bond valence sum to the
log
file. Using the same set parameters as in the
restrain example, set the BVS formula instead to an after
parameter.
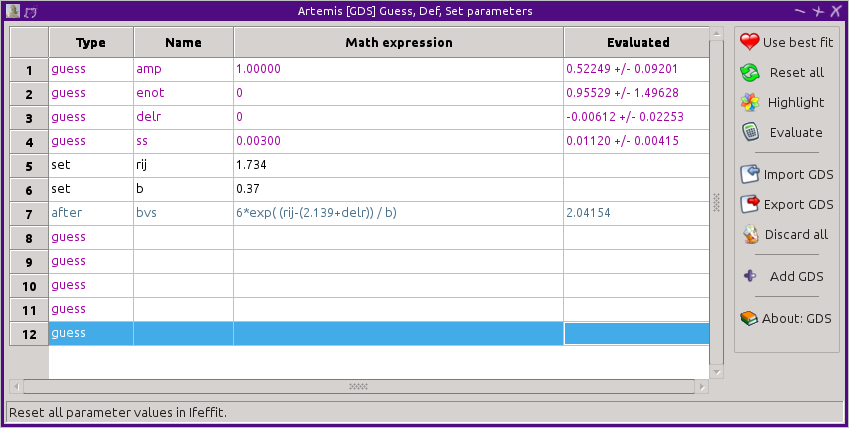
At the end of the fit, the BVS will be evaluated and reported in the
log file just below the guess, def, and set
parameters, like so:
after parameters:
bvs = 2.04154071 # [6*exp( (rij-(2.139+delr)) / b)]
![[Artemis logo]](../../images/Artemis_logo.jpg)