The HDF5 save file¶
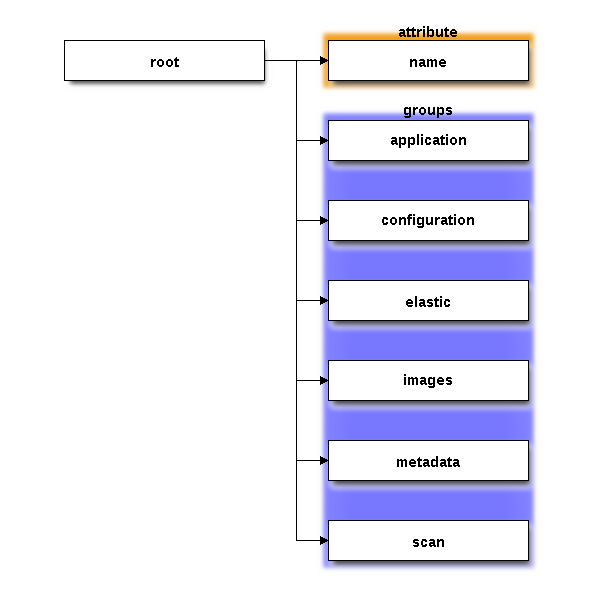
The top level structure of the save file looks like the diagram below. The root has a single attribute which identifies the creator of the HDF5 file. This is likely to be "Metis", although if we ever get around to packing the images into an HDF5 file at the beamline, then the creator attribute might be something else.
The “elastic” and “images” groups contain the actual content of the various images made during the measurement. The “elastic” group contains the sequence of elastic images, while the “images” groups contains the set of resonant or non-resonant XES images.
The other groups are a way of organizing the many parameters of the METIS analysis project, including the various attributes of the Xray::BLA object, the XDI metadata, and the contents of the scan file.
The “application” group contains debugging information – mostly version numbers of major packages used by METIS, a string identifying the computer platform used to create the file, and a timestamp.
| Name | Description |
|---|---|
| root | |
| name | name of program creating file |
| application | Version numbers, timestamp |
| configuration | Xray::BLA attributes |
| elastic | collection of elastic images |
| images | collection of XES images |
| metadata | XDI metadata |
| scan | information from scan file |
The elastic group¶
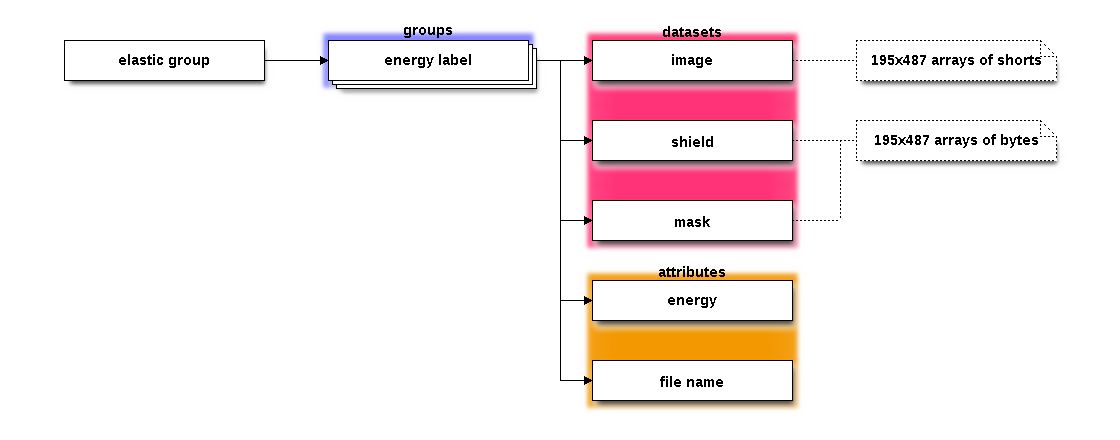
The elastic group contains the sequence of elastic images, each image
in a subgroup in the group. The name of the data set is the energy
portion of the filename of the measured tiff image. In the case of
one of the Nb Kβ2,4 measurements seen throughout this
manual, the filename of an elastic image might be something like
NbF5_Kb2_elastic_189555_0001.tif. That image was measured
with the incident energy at 18955.5 eV. The decimal was removed to
make the filename. The name of the group containing this image is
189555.
The subgroups each contain three datasets. These datasets contain the
original image (image), the shield if used (shield), and the
final mask (mask).
Each subgroup has two attributes associated with it, called energy
and filename. In the case of dataset 189555, the energy
attribute is set to the energy value, 18955.5 eV, and the filename
is set to the fully resolved filename of the tiff file.
The images group¶
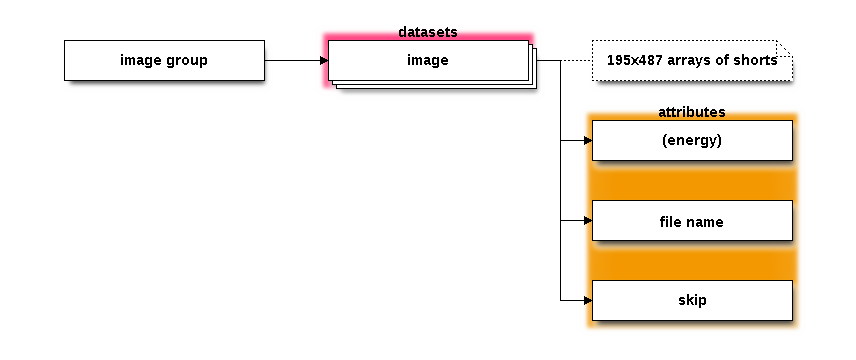
The images group contains all of the resonant or non-resonant XES
images made as part of the measurement. In XES mode, this will
typically be some number of repetitions made at an energy well above
the edge. In HERFD mode, this will be sequence of images made at
each point in the XANES scan. In RXES and Mask modes, this
group will be empty.
In HERFD mode, each dataset in this group will have an energy
attribute giving it's energy in the XANES scan. In XES mode, the
energy attribute is absent. The filename attribute is set to
the fully resolved filename of the tiff file. The skip attribute
is a flag which tells METIS whether to exclude an image
from the analysis. For example, in XES mode, setting this to a
false value for an image would exclude it from a merge of the
resulting XES spectra. In HERFD mode it would be excluded from
the XANES spectrum (not unlike deglitching).
The configuration group¶
This group has no datasets, just a lot of attributes for capturing much of the structure of the Xray::BLA object in the context of METIS, this group captures the values of most of the controls on the Files and Mask pages as well as all of the contents of the Configuration page.
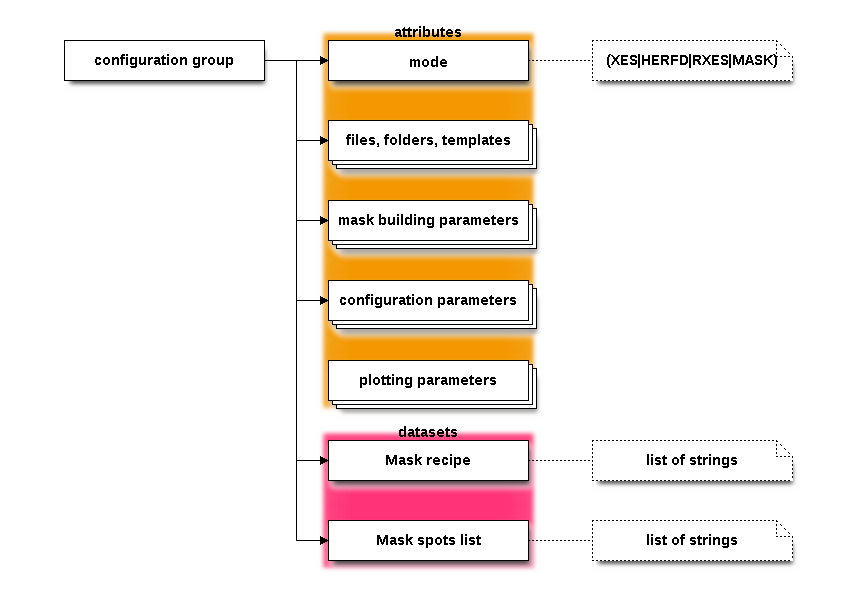
Use the configuration/mode attribute to determine what
METIS mode this HDF5 file was created in.
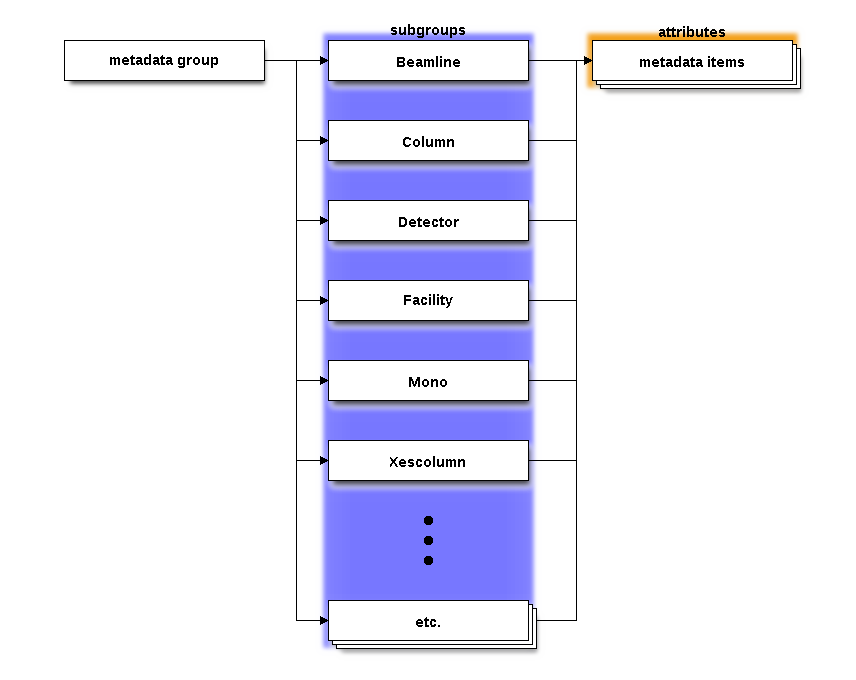
The metadata group¶
This group has no datasets, just a bunch of groups for the XDI metadata families. Each subgroup has a lot of attributes which capture everything from the XDI page.
There is a subgroup for each of the defined semantic groupings
used in an XES measurement. There is another subgroup called
Xescolumns which is used when exporting a column data file
containing an XES spectrum. Any other metadata families defined by
the user will be exported into their own subgroups.
Todo
BLA and Pilatus metadata families
The scan group¶
The scan file is simply slurped into the HDF5 file and stored as the
contents attribute of the scan group.
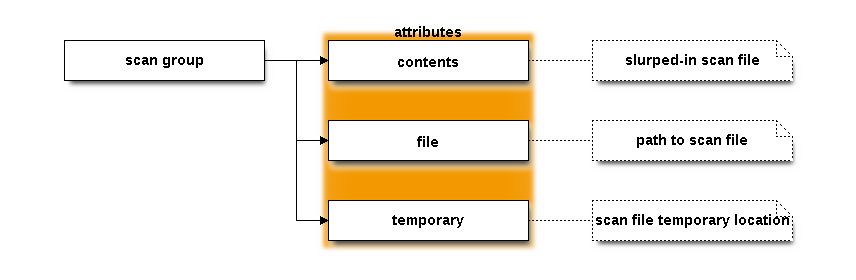
The file attribute contains the full path to the slurped-in file.
The temporary attribute holds the full path to the location where
METIS writes out the stash file temporarily for use in
HERFD and RXES modes. That changes between instances of the
program.
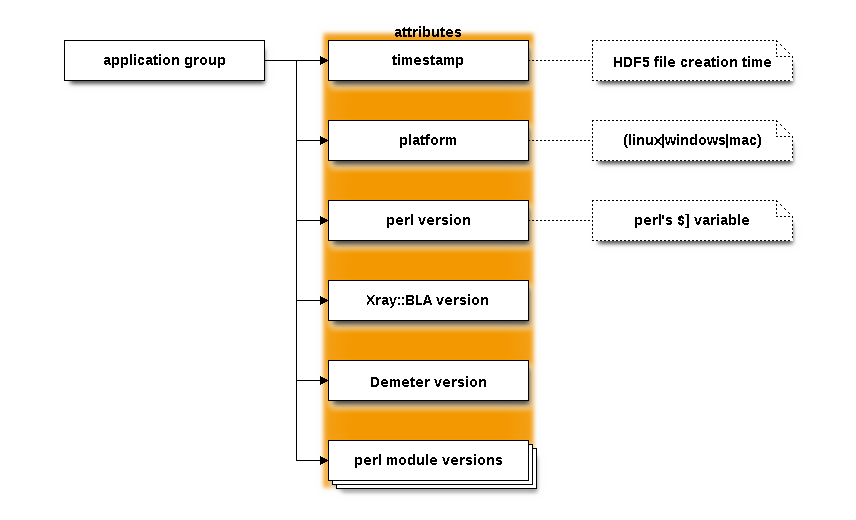
The application group¶
This groups contains attributes explaining the state of the program, including a timestamp, the platform on which it was run, and the version numbers of many of the software components. This is mostly useful for diagnostic purposes.
Xray::BLA and METIS are copyright © 2011-2014, 2016 Bruce Ravel and Jeremy Kropf – This document is copyright © 2016 Bruce Ravel
This document is licensed under The Creative Commons Attribution-ShareAlike License.
If this software and its documentation are useful to you, please consider supporting The Creative Commons.
